Superinfection exclusion
Alpha
Beta
Gamma
Delta
The majority of readers will identify these as the current circulating variant strains of Covid {{1}}. The World Health Organisation decided upon this naming system as being easier and more practical to discussed (sic) by non-scientific audiences {{2}}.
All viruses vary, and viruses with genomes made from ribonucleic acid (RNA) vary more than those which have deoxyribonucleic acid (DNA) genomes. This is because the enzymes that replicate RNA virus genomes do not have an error correction facility.
The virus that causes Covid-19 is an RNA virus and so is Deformed wing virus (DWV), probably the most significant virus to bees and beekeepers (other than those who have Covid that is).
Virus variation
Why does virus variation matter?
Or, asking the same question in a more roundabout way, why don’t RNA viruses evolve error correcting enzymes (after all, the DNA viruses have these … can it be that difficult?).
If the enzyme makes errors then that’s surely a bad thing?
Actually … for the particular ‘lifestyle’ that these viruses practice, errors and variation are a good thing.
They benefit the virus.
But you know all this already, even if you don’t think you do.
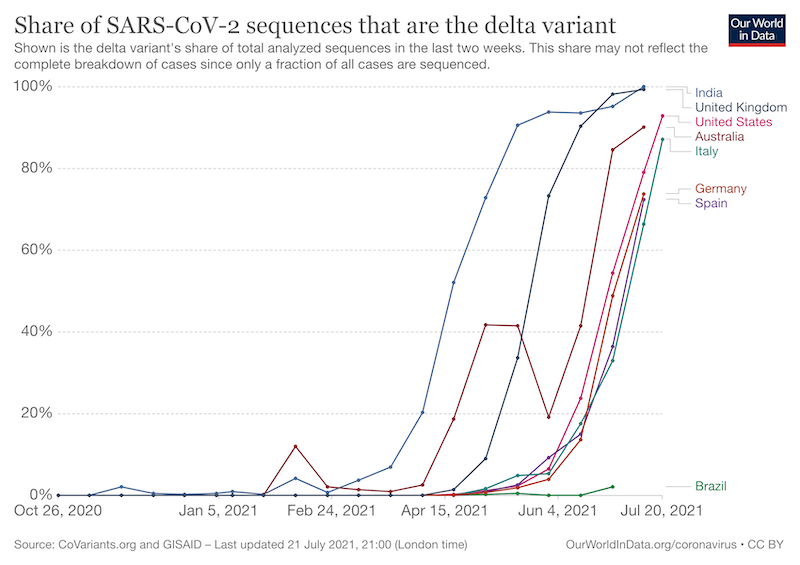
The SARS-Cov2 delta variant now accounts for at least 99% of cases of Covid-19 in the UK. It accounted for just 0.1% of cases in late February.
The delta variant is much more transmissible. It carries errors, or mutations as they’re more correctly known, that – for whatever reason – means it can be passed from person to person much more efficiently {{3}}.
These mutations benefit the virus and allow it to spread further and faster {{4}}.
If the mutations (‘errors’) that the virus acquires are beneficial – by increasing transmission, by expanding the cell, tissue or host range, by helping evade the immune response in a partially vaccinated population (!) for example – then the virus will successfully replicate and produce more viruses carrying the same mutation.
And a bunch of additional ones as well … acquired during the last round of replication.
Strains and types
At some point a virus acquires sufficient mutations from an earlier incarnation that it’s identified as a distinct strain.
For example, SARS-Cov2 is ~82% identical at the genome level to SARS-Cov1 which caused the SARS pandemic in 2003 {{5}}. SARS-Cov2 did not evolve from SARS-Cov1, but they share a common ancestor. They are different strains or types of coronavirus.
There are no hard and fast rules that define when a virus is considered a different strain or type. Often it’s historical, reflecting the geographic origin, or the source from which the virus was isolated. Different strains may exhibit different phenotypes – host range, transmission, disease etc. – but don’t have to.
For example (before we get back to honey bee viruses) there are three ‘types’ of poliovirus that are about 80% identical at the genetic level. They all cause exactly the same disease (poliomyelitis) and they replicate in an identical manner – same cell, tissue and host range for example. However, to the human immune system they ‘look’ different. The immune response to poliovirus type 1 will not protect you from infection with poliovirus type 3. That’s why the poliovirus vaccine contains a mixture of all three types, to protect you from all polioviruses.
You can even get infected with two types of poliovirus simultaneously, and the virus can replicate in the same cells in your gut … or, if you’re unfortunate, your brain.
I’ll return to dual infections shortly as it’s an important topic … and related to the study I’m going to discuss.
Deformed wing virus
There are two types of DWV, designated type A and type B {{6}}.
Originally these had names that reflected their original isolation.
DWV type A was termed Deformed wing virus and was isolated from honey bees displaying the characteristic symptoms of developmental deformities shown in the image below.
DWV type B was termed Varroa destructor virus type 1 and was isolated from the ectoparasitic mite Varroa destructor.
These viruses are very similar to each other. They are something like 85% identical at the level of the RNA genome. More importantly than this genetic identity (or perhaps similarity would be a better term to use here) is the fact that they appear to cause very similar diseases in honey bees.
Although early studies suggested there were some differences in their virulence, more recent work from Prof. Rob Paxton in Germany, from my lab, and from Dr. Eugene Ryabov and colleagues in the USA suggests these two types of DWV are actually very similar, at least in pathogenesis.
There do appear to be some differences, with the suggestion that type A does not replicate in Varroa whereas (full disclosure, the following study is from my lab) type B does. Undoubtedly, the other genetic differences between the types will confer some subtle variation in phenotype (effectively what they ‘do’), but – as far as beekeeping is concerned – they should probably be considered the same.
A protective, non-lethal type A DWV?
All of which made a 2015 colony-level study {{7}} of DWV infection rather intriguing.
This reported the survival of colonies that were infected with a “non-lethal” type B strain which were protected from infection with the “lethal” type A strain {{8}}. The authors summarised the significance of this study like this:
We propose that this novel stable host-pathogen relationship prevents the accumulation of lethal variants, suggesting that this interaction could be exploited for the development of an effective treatment that minimises colony losses in the future.
At the time there was a flurry of excitement and discussion about this {{9}}.
Superinfection exclusion
They proposed that the mechanism that prevented the infection with the type A strain was superinfection exclusion.
Virologists love mechanisms … 🙂
Which brings me back to virus variation.
Imagine a population of variant viruses trying to infect a new host … like a bee.
In mixed infections, a virus that has an advantage over the others in the population ends up ‘winning’ the competition for the resources of the host. They therefore make more progeny viruses.
One of the advantages could be that the virus simply replicates faster.
Another – more subtle, but the same outcome – is that the virus prevents other viruses from infecting the same cell (and, by extension, host).
By excluding competing viruses it effectively ‘wins’ the competition.
And some viruses do exactly this using a variety of cellular mechanisms.
For example, many viruses turn off the expression of the cellular receptor (think of this as the door) they use to enter the cell. If there’s no receptor (no door) then another virus cannot enter.
With no other viruses to compete with in the same cell there can only be one type of virus produced {{10}}.
There are other mechanisms as well, but we’ll stick with the receptor one as it’s easy to comprehend.
Superinfection of a cell containing a virus that has the ability to turn off the cellular receptor it uses will effectively exclude the second virus from replicating … hence superinfection exclusion.
What don’t we know about DWV?
A lot 🙁
We don’t know how it gets into cells. We don’t know a huge amount about its replication and we know precious little about the way it interacts with the cellular machinery (the ‘stuff’ in the cell that the virus hijacks to make more viruses) of the host.
However, since 2015 we do know that all the types of DWV that have been carefully studied appear to be more or less equally virulent. None appear to be ‘non-lethal’ as claimed for the type A virus in the superinfection exclusion paper.
This prompted us to look in a bit more detail at the consequences of dual or sequential infections with DWV in the laboratory.
Is there a precedence at work?
In mixed infections, does one virus always ‘win’ and predominate in the new virus population?
In sequential infections, does it matter the order in which the viruses are acquired?
And mixed infections are pretty much the norm for DWV infection. All bees, whether previously exposed to Varroa or in Varroa-free regions, appear to have low levels of DWV already present. If parasitised by the mite, these bees must experience a mixed infection.
In addition, studies we published several years ago {{11}} showed that recombinant viruses – essentially hybrids between type A and type B DWV – often predominated in heavily Varroa-infested colonies {{12}}.
If mixed infections cannot occur, how do such hybrids form?
Some of the answers to these questions are in our recently paper published in the ISME Journal. Gusachenko, O. et al., (2021) First come, first served: superinfection exclusion in Deformed wing virus is dependent upon sequence identity and not the order of virus acquisition. ISME J (2021). https://doi.org/10.1038/s41396-021-01043-4
Mixed DWV infections
I don’t propose to give a pupa-by-pupa account of the studies we conducted. You can read the paper – it’s open access and (because Olesya ‘Alex’ Gusachenko, the lead author, did most of the writing) relatively easy to comprehend 😉
But here are a few highlights.
Over the last few years we have produced reagents that allow us to produce almost ‘pure’ stocks of type A, type B or hybrid type A/B {{13}} strains of DWV.
At least 99.99% of these stocks are of one DWV type. Note that there will still be variation within this population as the replication errors probably generate one mutation per virus in the population. We therefore refer to these virus stocks as near clonal.
Injection of honey bee pupae with any of these viruses resulted in very similar levels and kinetics of replication – all the viruses replicate as far and as fast as each other.
In mixed injections, when two viruses were administered simultaneously, both replicated to equivalent levels.
We therefore found no evidence for the dominance of one strain over another.
Sequential DWV infections
But it got more interesting when we did sequential injections. We did these by injecting with one virus, waiting 24 hours and then injecting with a second virus.
Using type A and type B DWV both viruses had replicated to similar high levels (billions of virus per pupa) within 48 hours, irrespective of the order of addition.
If superinfection exclusion was operating we would have perhaps expected type A to have prevented or reduced the replication of type B. However, that didn’t appear to be the case.
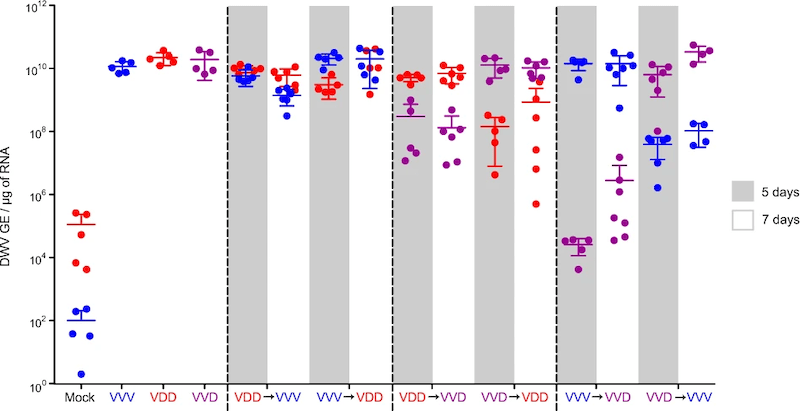
Competition between sequential infecting DWV isolates. VVV is type B, VDD is type A and VVD is a hybrid between them.
But, when we looked at sequential infections between type A and a type A/B hybrid we did see that replication of the second virus was delayed.
Delayed, but not stopped altogether.
It would take a complete post to describe the figure above 🙁 . We’ve quantified the virus present 5-7 days after sequential injections with type A (VDD), type B (VVV) or a hybrid virus (VVD) {{14}}.
The columns labelled VDD→VVV or VVV→VDD show the viruses and order of addition. The dots represent the amount of virus present at 5 or 7 days post injection. When the viruses were more similar to each other – for example, the VVV→VVD or VVD→VVV pairs on the right – there was a greater impact on the replication of the virus added second.
The same but different
We extended these studies to look at sequential infections with two viruses that only differed by 4 nucleotides (the building blocks) of the 10,140 nucleotides in the RNA genome of DWV i.e. 99.6% identical.
Cunningly, these four differences allowed us to unambiguously identify which virus was replicating.
In this part of the study the virus added second did not replicate to detectable levels.
So … our data clearly demonstrates that viruses that were more similar to each other were more likely to inhibit replication during sequential infections.
In addition, no individual virus type was dominant over any others.
This didn’t look much like classic superinfection exclusion to us.
Red or green viruses
Not content with generating graphs and tables we went on to take photographs of virus infected pupae.
You can’t beat a nice colour image when trying to impress the peer reviewers 😉 .
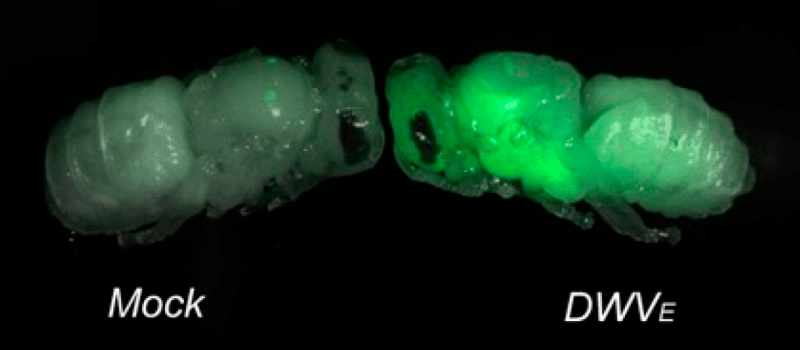
Remember that DWV is too small to see with even the most powerful light microscope. You could fit several billion on the head of a pin.
We therefore engineered the virus genome to ‘show’ us where it was replicating.
We did this by introducing an additional gene that fluoresced green or red when under UV light. I’ve discussed green viruses before … the red version uses similar technology, but using a different fluorescent reporter gene.
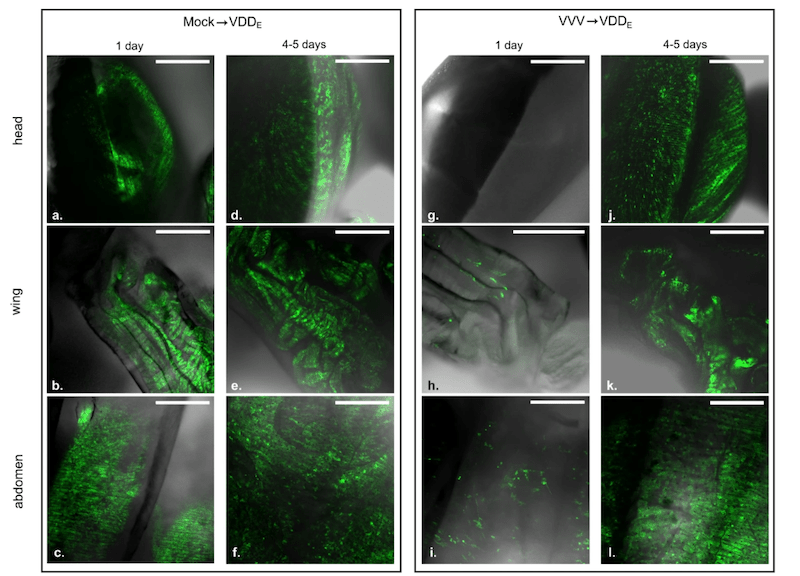
DWV replication (showed by green fluorescent signal) in the head, wing and abdomen of honey bee pupae
Using the red or green viruses we showed very similar results to those described above. When we superinfected with a genetically similar virus, its replication was inhibited. When it was genetically more divergent it could replicate (and we could visualise it as red or green foci of infection in a variety of tissues of the developing pupa).
Red and green viruses
We also infected bee with the red and green viruses simultaneously. Most of the fluorescent foci of infection were red or green, but a small number were both red and green.
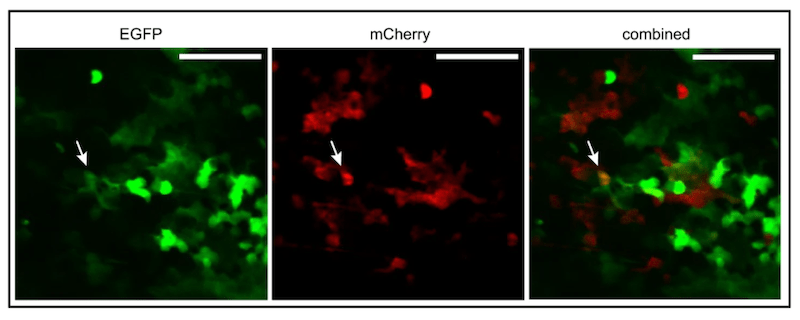
Green (EGFP) and red (mCherry) expressing DWV coinfecting a honey bee pupa. Arrow indicates dually infected cells.
The most likely explanation for having both colours overlapping in the photograph was because the virus were replicating in the same cells in the honey bee pupa.
Since this was exactly the sort of situation that was needed to generate recombinants (hybrids or chimeras) between the two different DWV viruses we specifically looked for them {{15}}.
And there were lots and lots of recombinants …
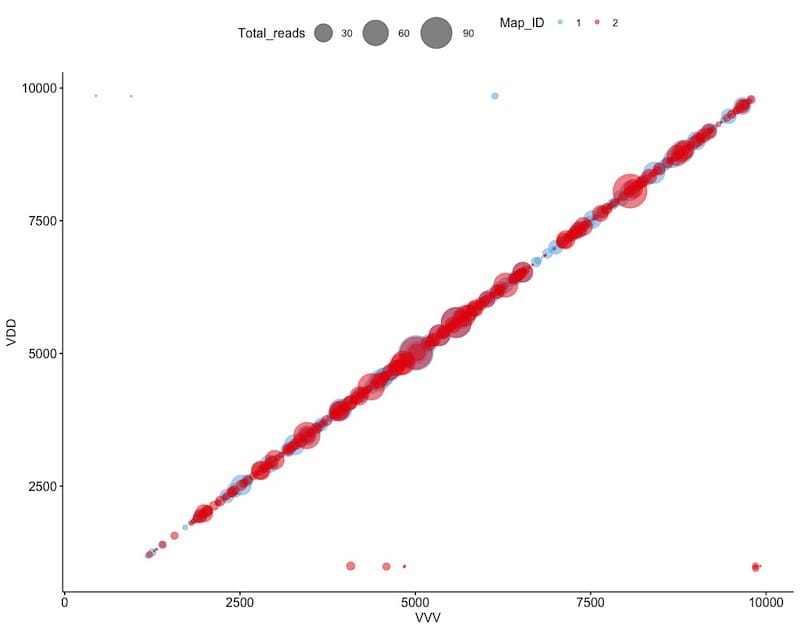
Recombination between DWV viruses. The size and position of ‘bubbles’ indicate the location and number of junctions.
The bubble plot above shows the location and frequency of junctions. One virus is plotted on the horizontal and one on the vertical axis. It’s a sort of two-dimensional map of the virus. Think of a junction as where one virus ends and the other starts. They are located throughout the DWV genome – hundreds of them.
This suggests that pupae infected with both type A and type B DWV will act as ‘factories’ for the production of thousands more different hybrid variants between the two viruses.
Most of these hybrids will grow poorly.
Many will be uncompetitive.
But some – like the delta variant of SARS-Cov2 – might be more transmissible.
And some could be more pathogenic.
Or – the nightmare scenario – both 😯 .
What’s this got to do with practical beekeeping?
Every time a beekeeper moves bees about s/he is also moving viruses about.
This happens when you move bees to out apiary, when selling a nuc or when importing a queen.
This will contribute to the constant mixing of DWV variants that occurs when bees drift between hives, when drones mate with queens, when phoretic Varroa jumps onto a bee that is robbing a collapsing colony.
There’s a difference of course.
All those bee-driven mixing events are local and small scale … a few bees and a few miles.
But if you import a nuc from Greece via Northern Ireland both the distance and number of bees (and hence number of viruses) is much greater.
Of course, most of this mixing will just generate more mixtures of viruses.
It will also generate more recombinants.
But there’s always the possibility it might throw up a highly virulent, highly transmissible variant.
Which would not be a ‘good thing’.
And if it does, a ‘non-lethal type A strain’ (should such a thing actually exist) is not going to help prevent infection by the mechanism of superinfection exclusion I’m afraid 🙁 .
Without doubt the best way to prevent infection is to minimise the mite numbers in your colonies. This is a subject I’ll be tackling again in a couple of weeks.
But, before I go, do we understand how the more closely related strains of DWV prevent superinfection?
Yes … probably, and it’s all to do with the immune response of the bee.
I’ll discuss this in the future as it’s a mechanism that could be exploited to produce bees immune to the ravages of DWV.
{{1}}: If you’ll excuse an early diversion into pedantry, the disease Covid-19 is caused by the virus SARS-Cov2.
{{2}}: With no mention of avoiding causing offence to the good people of Kent, South Africa, Brazil and India, where these viruses first appeared. Ebola virus is still called Ebola virus … Peter Piot, who discovered and named the virus after the river in DRC, was well aware that named associations can have negative connotations. Ebola virus was actually discovered about 75 miles away in the town of Yambuku.
{{3}}: Remember there are a further 20 letters in the Greek alphabet … once we get to omega, like car number plates, they’ll have to come up with a new numbering scheme.
{{4}}: And, I fear, we’re going to see just how far and how fast over the next 8-12 weeks.
{{5}}: There were only ~8000 cases of SARS in 2003 but the case fatality rate was 10%.
{{6}}: As an aside, a third DWV type has been reported – imaginatively designated type C – though it might be restricted to Devon as there have been no further reports of it elsewhere to my knowledge.
{{7}}: Mordecai, G., Brettell, L., Martin, S. et al. Superinfection exclusion and the long-term survival of honey bees in Varroa-infested colonies. ISME J 10, 1182–1191 (2016). https://doi.org/10.1038/ismej.2015.186
{{8}}: It should be noted that this was published before many of the studies showing how similar the type A and B strains were in their pathogenesis.
{{9}}: At least amongst a small group of geeky bee virologists and scientifically-interested beekeepers.
{{10}}: Or, more pedantically correctly, one type of virus plus the inherent variation generated when these viruses replicate.
{{11}}: Moore J, Jironkin A, Chandler D, Burroughs N, Evans DJ, Ryabov EV. Recombinants between Deformed wing virus and Varroa destructor virus-1 may prevail in Varroa destructor-infested honeybee colonies. J Gen Virol. 2011 92:156-61. doi: 10.1099/vir.0.025965-0.
{{12}}: Ryabov EV, Wood GR, Fannon JM, Moore JD, Bull JC, Chandler D, et al. (2014) A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees (Apis mellifera) Prevails after Varroa destructor-Mediated, or In Vitro, Transmission. PLoS Pathog 10: e1004230. https://doi.org/10.1371/journal.ppat.1004230.
{{13}}: Or type B/A which is the same thing, but the other way round.
{{14}}: Yes, the naming scheme is a bit messy but there are good historical reasons for it. Unlike the WHO, we don’t have the luxury of renaming these viruses … if we did, other scientists would struggle to understand our published results.
{{15}}: My lab have a long-standing interest in virus recombination and have published a series of papers on the molecular mechanism by which it occurs. An absolute requirement is that both viruses are present in the same part of the same cell.


Join the discussion ...